Compound: PYROJACAREUBINE_COMPOUND-3
Project: MOLECULES,15,7106[2010]CSEARCH-NUMBER:UWLU072496-20131004

Did you know ? |
You can donate your private spectral data to the 'community'. |
|
Request from: [email protected] Compound: PYROJACAREUBINE_COMPOUND-3 Project: MOLECULES,15,7106[2010]CSEARCH-NUMBER:UWLU072496-20131004 |
 |
|
Compound: PYROJACAREUBINE_COMPOUND-3 Project: MOLECULES,15,7106[2010]CSEARCH-NUMBER:UWLU072496-20131004 | 
|



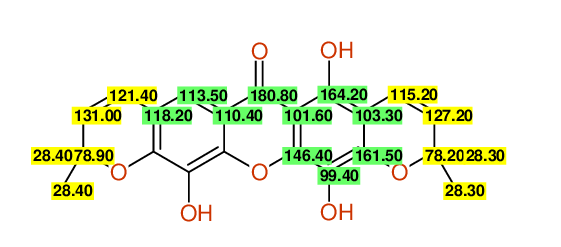
| Carbon number | Chemical Shift Value | Multiplicity from Structure | Multiplicity from Experiment |
| 1 | 164.20 | S | - |
| 2 | 103.30 | S | - |
| 3 | 161.50 | S | - |
| 4 | 99.40 | S | - |
| 5 | 146.40 | S | - |
| 9 | 118.20 | S | - |
| 10 | 113.50 | D | - |
| 11 | 110.40 | S | - |
| 12 | 180.80 | S | - |
| 13 | 101.60 | S | - |


| Carbon number | Chemical Shift Value | Exchange Flag | Multiplicity from Structure | Multiplicity from Experiment |
| 14 | 121.40 | A | D | - |
| 15 | 131.00 | B | D | - |
| 16 | 78.90 | C | S | - |
| 17 | 28.40 | D | Q | - |
| 18 | 28.40 | D | Q | - |
| 19 | 115.20 | A | D | - |
| 20 | 127.20 | B | D | - |
| 21 | 78.20 | C | S | - |
| 22 | 28.30 | D | Q | - |
| 23 | 28.30 | D | Q | - |






| Checking lines & multiplicity | Carbons/Lines | Singlet | Dublet | Triplet | Quartet | Odd | Even | None |
| From structure | 23 | 14 | 5 | 0 | 4 | 14 | 9 | 0 |
| From spectrum | 20 | 11 | 5 | 0 | 4 | 11 | 9 | 0 |

| Flag | Number of Shifts | Lowest Shift value | Highest Shift value | Range | Multiplicity | Comment |
| A | 2 | 115.20ppm | 121.40ppm | 6.20ppm | D |
Range is large - please check Unflagged line within range ( 118.20 ppm ) Unflagged line less than 2.0ppm away from exchange-group ( 113.50 ppm ) |
| B | 2 | 127.20ppm | 131.00ppm | 3.80ppm | D | |
| C | 2 | 78.20ppm | 78.90ppm | 0.70ppm | S | |
| D | 4 | 28.30ppm | 28.40ppm | 0.10ppm | Q | |

|
|

| Carbon Number | Neural Network Prediction |
HOSE-Code Prediction |
Preferred Value from both Predictions |
Experimental values |
Difference (Exp-Pred/ppm) |
Assignment | Prediction Quality |
|---|---|---|---|---|---|---|---|
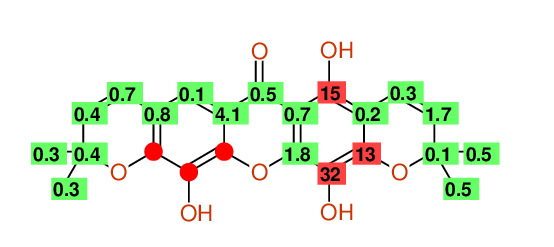
| 1 | 150.5 | 148.7 | 149.1 | 164.2 | 15.1 | Assigned by author | Only very few similar structures |
| 2 | 104.9 | 103.5 | 103.5 | 103.3 | 0.2 | Assigned by author | |
| 3 | 146.3 | 148.8 | 148.3 | 161.5 | 13.2 | Assigned by author | Only very few similar structures |
| 4 | 128.7 | 134.0 | 131.4 | 99.4 | 32.0 | Assigned by author |
Large Difference between NET & HOSE Only very few similar structures Inconsistent assignments in reference material |
| 5 | 143.9 | 145.3 | 144.6 | 146.4 | 1.8 | Assigned by author | |
| 6 | 145.0 | 144.1 | 144.5 | Chemical shift not available | |||
| 7 | 136.6 | 135.1 | 135.4 | Chemical shift not available | |||
| 8 | 146.2 | 146.4 | 146.4 | Chemical shift not available | |||
| 9 | 126.3 | 117.4 | 117.4 | 118.2 | 0.8 | Assigned by author |
Large Difference between NET & HOSE Inconsistent assignments in reference material |
| 10 | 115.0 | 113.4 | 113.4 | 113.5 | 0.1 | Assigned by author | |
| 11 | 117.3 | 113.9 | 114.5 | 110.4 | 4.1 | Assigned by author | |
| 12 | 180.9 | 181.7 | 181.3 | 180.8 | 0.5 | Assigned by author | |
| 13 | 99.5 | 102.3 | 100.9 | 101.6 | 0.7 | Assigned by author | Only very few similar structures |
| 14 | 126.4 | 120.7 | 120.7 | 121.4 | 0.7 | Exchangeable assigned by author |
Large Difference between NET & HOSE Inconsistent assignments in reference material |
| 15 | 129.3 | 130.6 | 130.6 | 131.0 | 0.4 | Exchangeable assigned by author | |
| 16 | 78.2 | 78.5 | 78.5 | 78.9 | 0.4 | Exchangeable assigned by author | |
| 17 | 27.8 | 28.1 | 28.1 | 28.4 | 0.3 | Exchangeable assigned by author | |
| 18 | 27.8 | 28.1 | 28.1 | 28.4 | 0.3 | Exchangeable assigned by author | |
| 19 | 120.3 | 115.5 | 115.5 | 115.2 | 0.3 | Exchangeable assigned by author | |
| 20 | 129.1 | 128.9 | 128.9 | 127.2 | 1.7 | Exchangeable assigned by author | |
| 21 | 78.2 | 78.1 | 78.1 | 78.2 | 0.1 | Exchangeable assigned by author | |
| 22 | 27.8 | 27.8 | 27.8 | 28.3 | 0.5 | Exchangeable assigned by author | |
| 23 | 27.8 | 27.8 | 27.8 | 28.3 | 0.5 | Exchangeable assigned by author |






| HOSE | NET | NET&HOSE | NONE |









| ||
 |
 |

|

Compound: PYROJACAREUBINE_COMPOUND-3 |

|
| Experimental values | ||||
 | ||||
| Symmetry considerations | ||||
 | ||||
| Predicted values | ||||
 | ||||
| Matching map | ||||
 | ||||
| Deviation per position ( Average is 3.7ppm ) | ||||
 | ||||
 | ||||
| ||||
 | ||||
| Overall Similarity Index is 9.5 0.0 is a "perfect match", up to approximately 3.0 it is "reasonable", above 5.0 it is more or less "unbelievable" |

| ||
 |
 |
685 Requests have been launched by [email protected] | |||||
Year |
Accept |
Minor Revision |
Major Revision |
Reject | Only Prediction |
| 2021 | 36 | 453 | 165 | 31 | |